One day in January 1976,
a young venture capitalist and a microbiology professor walked into a bar near
the University of California, San Francisco (UCSF), and the originally
scheduled ten minute meeting was extended to three hours. From that moment on, a
company that changed the history of biotechnology was born. They named the
company Genentech with the abbreviation of genetic engineering technology.

Figure 1. Dr. Herbert Boyer, co-founder of Genentech (left) and venture capitalist Robert a.swanson (right) (source: data figure)
Genentech successfully
recombined the DNA of human insulin into the plasmid in E. coli cells for the
first time, and let E. coli as a cell factory to produce recombinant human
insulin and commercialize it. Since then, genetic engineering has officially
kicked off.
However, after more than 40 years, people have recombined and expressed insulin at the technical level, and almost only done one thing - put the exogenous DNA into the cell, let the gene be transcribed into RNA, translated into protein, and then make some engineering transformation around the expression and translation efficiency of the exogenous DNA.
The efficient expression of proteins in host cells is not only information transmission. Raw material supply, peptide chain extension, post-translational modification, folding, secretion and even emergency repair may affect the efficiency of protein expression.
"To make the
delicate protein '3D printer' of cells work efficiently, a large number of
genes play an indispensable role at the genome-wide level. However, although we
recognize this at the level of basic research, at the level of engineering, it
is still difficult to achieve genome-wide engineering to improve the protein
expression efficiency of cells, and our current understanding is still very
shallow." Professor Zhang Zhen of Tsinghua University said.

Figure 1 Zhang Zhen
So far, cell factories
have been able to produce antibiotics, amino acids, recombinant proteins,
bioenergy, bioplastics and even "artificial meat", which are widely
used in biological manufacturing, pharmaceutical, food, energy, agriculture and
other fields.
However, like the case of recombinant insulin synthesis, at present, people have more modifications to the external source pathway, but less understanding of the chassis cell itself at the genome-wide level, which restricts the ability of its systematic engineering transformation, and the potential of the cell chassis itself has not been systematically explored.
If the gene sequence of
the exogenous pathway is compared to the drawing and the cell to the workshop,
most of the existing efforts are focused on the "drawing", but they
still lack the ability to systematically recognize the overall situation of the
"workshop" and engineering transformation.
From random mutagenesis to genome-wide customization, a number of technologies have given birth to discover chassis cells
The core of the
construction of synthetic biology cell factory is how to get the desired
industrial phenotype by designing the appropriate genotype. Professor Zhang
Zhen believes that in the era of genome, scientists can get a large number of
genotype related test data through various public biological databases, but
what is really valuable is to get genotype data related to industrial
phenotypes.
Figure industrial phenotypes often concerned by synthetic biology (source: the team)
With the continuous
development of molecular biology and genetic engineering research methods, the
construction strategy of cell factory has experienced different historical
stages. Compared with the early method of obtaining high-yield strains of
target products mainly through irrational mutation breeding technology, since
the 1990s, with the gradual introduction of molecular biology and genetic
engineering technology, the discipline of metabolic engineering has been
officially established.
Metabolic engineering
uses recombinant DNA technology to purposefully design known metabolic pathways
in organisms, regulate and optimize gene networks in cells, and build cell
factories with specific functions, such as improving the yield of target
products.
However, most of the
design methods of metabolic engineering guidance are based on known biological
knowledge, because there are many unknown factors in the microbial metabolic
network that may affect the industrial phenotype of the target product, or
"dark matter of life". The efficiency of acquiring new knowledge by
this means is not high, and the process of cell factory transformation still
requires a lot of time and energy.
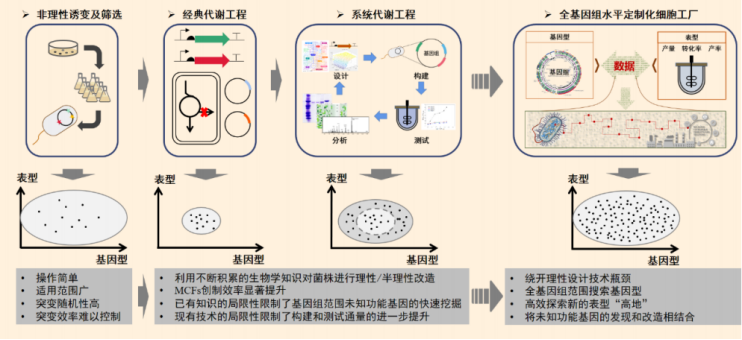
Figure 1 development process and Prospect of cell factory design and construction (source: the team)
So, in order to make the design of cell factories more efficient, how to analyze these "dark matter of life"?
"Through its own
technology platform, we can study the relationship between specific industrial
phenotypes and genotypes of microorganisms at the whole genome level in
parallel, so as to obtain a large-scale genotype industrial phenotype
associations (GPA) data set." Zhang Zhen said.
"In recent years, the reduction of the cost of high-throughput DNA synthesis, the leap of gene editing technology and second-generation sequencing technology, the maturity of high-throughput detection technology and many other technologies have made it possible to mine large-scale GPA data sets. These newly mined data will become the 'new continent' at the genome level." He said.
chassis cells refers to the newly discovered gene loci associated with the target industrial
phenotype. For example, they found many unexpected loci related to protein
synthesis, such as oxidative stress, which has positive help for protein
synthesis.
"After acquiring new knowledge and verifying its
industrial value based on the genome-wide scale association map, the system
engineering transformation on the chassis will provide a new path of 'discovery
engineering' for the system to improve the efficiency of the cell
factory." Zhang Zhen said.
Through the
"trilogy" of ultra-high throughput, fast and low-cost technology, we
try to combine genome-wide genotype and industrial phenotype .
Zhang Zhen's team has matured to build a set of technical platforms through which GPA data sets can be mined on a genome-wide scale with ultra-high throughput, speed and low cost.
The platform is supported
by three core technologies: CRISPR whole genome editing technology, ultra-high
throughput droplet microfluidic monoclonal culture and screening integration
technology and synthetic biosensor technology.
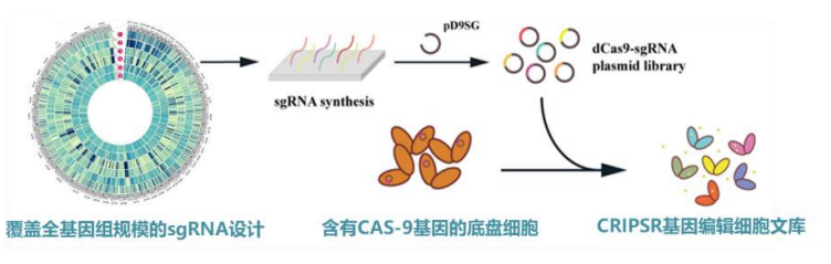
Figure genome wide CRISPR gene interference library (source: the team)
First, CRISPR genome-wide editing technology. The team has established a CRISPR gene interference library with a cell genome-wide scale of up to one million for typical industrial hosts. "Interference" here refers to gene knockdown or activation of chassis cells, and even gene editing [1,2,3].
Zhang Zhen said that the "high version" of genotype mutation was achieved through CRISPR editing technology on this platform. The "high version" here is because it has two characteristics: customizable and traceable. In other words, scientists can design interference or editing of sgRNA at any site, and after phenotypic changes, they can track the specific location of sgRNA without measuring the whole genome.
It is reported that at present, the team has a variety of mature industrial chassis cell genome-wide editing cell libraries, and has independently developed genome-wide sgRNA library design software and web applications.
Figure 1 development process of software tool for genome-wide sRNA library design (source: the team)
Second, we independently developed the integrated technology of droplet microfluidic cell culture and screening with million flux level screening ability. The "microbial droplet culture technology" developed by Zhang Zhen's team in combination with microfluidic technology, photoelectric sensing and control and automation technology can realize parallel culture, growth curve measurement and adaptive evolution of microbial droplets in various volume scales of skin lift, nano lift and micro upgrade.
The platform adopts integrated monoclonal culture, and the number of single monoclonal can exceed 106. Compared with the traditional method, the culture cost is reduced by about 1000 times. Moreover, the platform can automatically change the liquid, and the cell growth state is highly uniform, which is suitable for the growth of a variety of mature industrial microorganisms [4].
Zhang Zhen pointed out that through the control of environmental conditions, microorganisms in droplets may achieve "industrial similarity" culture. "This is equivalent to realizing the transformation of the independent genotype of the library into an independent reactor, allowing it to grow an accessible target phenotype."
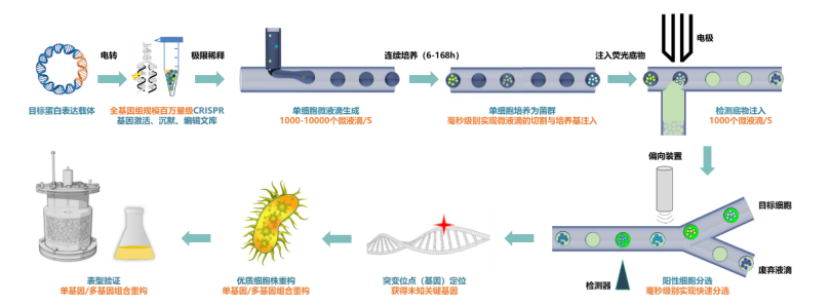
Figure integrated platform for ultra high flux droplet microfluidic monoclonal culture and screening (source: the team)
Third, the self-developed synthetic biosensor with high sensitivity and millisecond response. Usually, directional optical technology, especially fluorescence technology, will be used to test the phenotype of millions of skin upgrading droplets. To this end, the team established a series of synthetic biological fluorescence sensing technologies for quantitative testing of protein and small molecule concentrations [5,6,7].
Through this technology, the concentration of target molecules can be quantitatively converted into fluorescent signals. It has high sensitivity and fast response speed, and is fully compatible with the droplet microfluidic system with a million magnitude flux, laying the foundation for the mapping of the phenotype genotype correlation map of target metabolites.
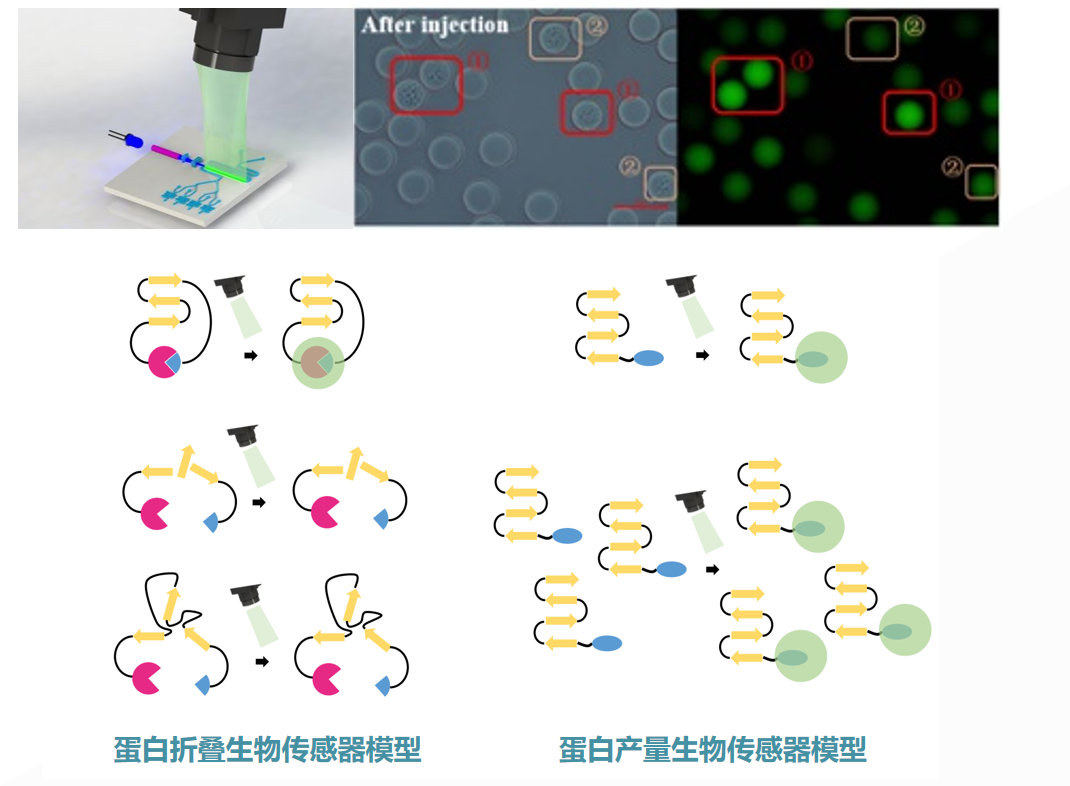
Figure: protein production and folding biosensor model (source: the team)
A centralized laboratory integrating dry and wet, integrating data islands, and creating the first open source enabling platform for synthetic biological data in China
At present, the databases used in the field of Bioscience, such as NCBI, KEGG, PDB, etc., are formed by the integration of discrete data supplied by the team of scientists, and they are mainly databases of scientific attributes. "Imagine that if we have a massive GPA database driven by industrial demand, we will have the core raw data for designing efficient cell factories." Zhang Zhen said.
He pointed out that from the perspective of the industry, although synthetic biology has great potential, the technology has developed very early, and there are still many problems to be solved. Synthetic biology is an interdisciplinary subject with both scientific and engineering attributes, but at this stage, scientific attributes are still heavy and engineering is weak. Especially in academia, many research and development methods are still in the "manual workshop" stage.
Zhang Zhen believes that through automation and high-throughput technology, the process of scientists doing experiments will become a central platform, so as to promote the transfer of synthetic biology from scientific attributes to engineering attributes. Once this chain is broken through, synthetic biology in the future will become a problem of pure information science and data science.
In the field of biological manufacturing, cell factory design is the development trend in the future. From the current discrete personal laboratory to the centralized laboratory platform, from distributed data to integrated large-scale data production, this highly standardized and high-quality data provides a great possibility for the final evolution to AI driven design.
If we can use large-scale GPA data sets to deeply mine unknown associated genes and their loci that cannot be found by traditional molecular biology methods from the whole genome based on data science methods, it will be possible to bypass the knowledge bottleneck of rational design from the perspective of data learning, and provide a new research paradigm for improving the efficiency of cell factory design and creation.
In addition, because the large-scale GPA data set has a wider search scope (genome-wide), and does not rely on existing knowledge, it will be possible to explore the phenotypic "highland" that previously could not be achieved by rationality / semi rationality, and obtain the next generation of customized cell factories with more efficient production efficiency and superior production performance.
For this platform, Zhang Zhen's team plans not only to provide a single point of service to scientists, but also to gradually implement academic open source to help the data standardization of multi-dimensional synthetic biology. Driven by high-throughput genotype industrial phenotype association map data, AI parses cell factories to create an enabling platform for synthetic biological data.
The platform will promote the upgrading of scientific research by engineering means, assist scientists to realize technological transformation, and connect the upstream and downstream of the industry. When the platform develops to a certain extent, it will accumulate high industrialization potential energy, and there will be a variety of industrialization possibilities in the future.
"After scientists confirm the strains and phenotypes they want to do by raising needs, we will help them do experiments, and then feed back the results to scientists. At the same time, we hope to deposit the data on the platform, and at the same time, we will open up academic contributions to obtain together, and verify its application value. In the future, this will become the core driving force for the design of several industrial phenotypes." Zhang Zhen said.
It is reported that the team has established extensive cooperation with relevant laboratories of key domestic universities, such as Tsinghua University, Tianjin University, East China University of technology, Shanghai Jiaotong University, Jiangnan University, Shanghai Institute of Botany, Chinese Academy of Sciences, etc. At the same time, Zhang Zhen is carrying out technology licensing and industrialization for the platform. At present, he is preparing to set up relevant companies and set up a technology research and development team. Professor Zhang Zhen is the chief scientist.
Zhang Zhen is full of expectations for the future development of the platform. He said: "I believe that having the core resources of data will also master the core information of cell factory design. We hope that such key databases will take root in China and serve China's local scientific research and industry."”
-End-
Article extracted from deeptech deep technology liuyakun
Original link:https://mp.weixin.qq.com/s/XzmtEshXYE2EvVehboDjAg
One day in January 1976,
a young venture capitalist and a microbiology professor walked into a bar near
the University of California, San Francisco (UCSF), and the originally
scheduled ten minute meeting was extended to three hours. From that moment on, a
company that changed the history of biotechnology was born. They named the
company Genentech with the abbreviation of genetic engineering technology.

Figure 1. Dr. Herbert Boyer, co-founder of Genentech (left) and venture capitalist Robert a.swanson (right) (source: data figure)
Genentech successfully
recombined the DNA of human insulin into the plasmid in E. coli cells for the
first time, and let E. coli as a cell factory to produce recombinant human
insulin and commercialize it. Since then, genetic engineering has officially
kicked off.
However, after more than 40 years, people have recombined and expressed insulin at the technical level, and almost only done one thing - put the exogenous DNA into the cell, let the gene be transcribed into RNA, translated into protein, and then make some engineering transformation around the expression and translation efficiency of the exogenous DNA.
The efficient expression of proteins in host cells is not only information transmission. Raw material supply, peptide chain extension, post-translational modification, folding, secretion and even emergency repair may affect the efficiency of protein expression.
"To make the
delicate protein '3D printer' of cells work efficiently, a large number of
genes play an indispensable role at the genome-wide level. However, although we
recognize this at the level of basic research, at the level of engineering, it
is still difficult to achieve genome-wide engineering to improve the protein
expression efficiency of cells, and our current understanding is still very
shallow." Professor Zhang Zhen of Tsinghua University said.

Figure 1 Zhang Zhen
So far, cell factories
have been able to produce antibiotics, amino acids, recombinant proteins,
bioenergy, bioplastics and even "artificial meat", which are widely
used in biological manufacturing, pharmaceutical, food, energy, agriculture and
other fields.
However, like the case of recombinant insulin synthesis, at present, people have more modifications to the external source pathway, but less understanding of the chassis cell itself at the genome-wide level, which restricts the ability of its systematic engineering transformation, and the potential of the cell chassis itself has not been systematically explored.
If the gene sequence of
the exogenous pathway is compared to the drawing and the cell to the workshop,
most of the existing efforts are focused on the "drawing", but they
still lack the ability to systematically recognize the overall situation of the
"workshop" and engineering transformation.
From random mutagenesis to genome-wide customization, a number of technologies have given birth to discover chassis cells
The core of the
construction of synthetic biology cell factory is how to get the desired
industrial phenotype by designing the appropriate genotype. Professor Zhang
Zhen believes that in the era of genome, scientists can get a large number of
genotype related test data through various public biological databases, but
what is really valuable is to get genotype data related to industrial
phenotypes.

Figure industrial phenotypes often concerned by synthetic biology (source: the team)
With the continuous
development of molecular biology and genetic engineering research methods, the
construction strategy of cell factory has experienced different historical
stages. Compared with the early method of obtaining high-yield strains of
target products mainly through irrational mutation breeding technology, since
the 1990s, with the gradual introduction of molecular biology and genetic
engineering technology, the discipline of metabolic engineering has been
officially established.
Metabolic engineering
uses recombinant DNA technology to purposefully design known metabolic pathways
in organisms, regulate and optimize gene networks in cells, and build cell
factories with specific functions, such as improving the yield of target
products.
However, most of the
design methods of metabolic engineering guidance are based on known biological
knowledge, because there are many unknown factors in the microbial metabolic
network that may affect the industrial phenotype of the target product, or
"dark matter of life". The efficiency of acquiring new knowledge by
this means is not high, and the process of cell factory transformation still
requires a lot of time and energy.

Figure 1 development process and Prospect of cell factory design and construction (source: the team)
So, in order to make the design of cell factories more efficient, how to analyze these "dark matter of life"?
"Through its own
technology platform, we can study the relationship between specific industrial
phenotypes and genotypes of microorganisms at the whole genome level in
parallel, so as to obtain a large-scale genotype industrial phenotype
associations (GPA) data set." Zhang Zhen said.
"In recent years, the reduction of the cost of high-throughput DNA synthesis, the leap of gene editing technology and second-generation sequencing technology, the maturity of high-throughput detection technology and many other technologies have made it possible to mine large-scale GPA data sets. These newly mined data will become the 'new continent' at the genome level." He said.
chassis cells refers to the newly discovered gene loci associated with the target industrial
phenotype. For example, they found many unexpected loci related to protein
synthesis, such as oxidative stress, which has positive help for protein
synthesis.
"After acquiring new knowledge and verifying its
industrial value based on the genome-wide scale association map, the system
engineering transformation on the chassis will provide a new path of 'discovery
engineering' for the system to improve the efficiency of the cell
factory." Zhang Zhen said.
Through the
"trilogy" of ultra-high throughput, fast and low-cost technology, we
try to combine genome-wide genotype and industrial phenotype .
Zhang Zhen's team has matured to build a set of technical platforms through which GPA data sets can be mined on a genome-wide scale with ultra-high throughput, speed and low cost.
The platform is supported
by three core technologies: CRISPR whole genome editing technology, ultra-high
throughput droplet microfluidic monoclonal culture and screening integration
technology and synthetic biosensor technology.

Figure genome wide CRISPR gene interference library (source: the team)
First, CRISPR genome-wide editing technology. The team has established a CRISPR gene interference library with a cell genome-wide scale of up to one million for typical industrial hosts. "Interference" here refers to gene knockdown or activation of chassis cells, and even gene editing [1,2,3].
Zhang Zhen said that the "high version" of genotype mutation was achieved through CRISPR editing technology on this platform. The "high version" here is because it has two characteristics: customizable and traceable. In other words, scientists can design interference or editing of sgRNA at any site, and after phenotypic changes, they can track the specific location of sgRNA without measuring the whole genome.
It is reported that at present, the team has a variety of mature industrial chassis cell genome-wide editing cell libraries, and has independently developed genome-wide sgRNA library design software and web applications.

Figure 1 development process of software tool for genome-wide sRNA library design (source: the team)
Second, we independently developed the integrated technology of droplet microfluidic cell culture and screening with million flux level screening ability. The "microbial droplet culture technology" developed by Zhang Zhen's team in combination with microfluidic technology, photoelectric sensing and control and automation technology can realize parallel culture, growth curve measurement and adaptive evolution of microbial droplets in various volume scales of skin lift, nano lift and micro upgrade.
The platform adopts integrated monoclonal culture, and the number of single monoclonal can exceed 106. Compared with the traditional method, the culture cost is reduced by about 1000 times. Moreover, the platform can automatically change the liquid, and the cell growth state is highly uniform, which is suitable for the growth of a variety of mature industrial microorganisms [4].
Zhang Zhen pointed out that through the control of environmental conditions, microorganisms in droplets may achieve "industrial similarity" culture. "This is equivalent to realizing the transformation of the independent genotype of the library into an independent reactor, allowing it to grow an accessible target phenotype."

Figure integrated platform for ultra high flux droplet microfluidic monoclonal culture and screening (source: the team)
Third, the self-developed synthetic biosensor with high sensitivity and millisecond response. Usually, directional optical technology, especially fluorescence technology, will be used to test the phenotype of millions of skin upgrading droplets. To this end, the team established a series of synthetic biological fluorescence sensing technologies for quantitative testing of protein and small molecule concentrations [5,6,7].
Through this technology, the concentration of target molecules can be quantitatively converted into fluorescent signals. It has high sensitivity and fast response speed, and is fully compatible with the droplet microfluidic system with a million magnitude flux, laying the foundation for the mapping of the phenotype genotype correlation map of target metabolites.

Figure: protein production and folding biosensor model (source: the team)
A centralized laboratory integrating dry and wet, integrating data islands, and creating the first open source enabling platform for synthetic biological data in China
At present, the databases used in the field of Bioscience, such as NCBI, KEGG, PDB, etc., are formed by the integration of discrete data supplied by the team of scientists, and they are mainly databases of scientific attributes. "Imagine that if we have a massive GPA database driven by industrial demand, we will have the core raw data for designing efficient cell factories." Zhang Zhen said.
He pointed out that from the perspective of the industry, although synthetic biology has great potential, the technology has developed very early, and there are still many problems to be solved. Synthetic biology is an interdisciplinary subject with both scientific and engineering attributes, but at this stage, scientific attributes are still heavy and engineering is weak. Especially in academia, many research and development methods are still in the "manual workshop" stage.
Zhang Zhen believes that through automation and high-throughput technology, the process of scientists doing experiments will become a central platform, so as to promote the transfer of synthetic biology from scientific attributes to engineering attributes. Once this chain is broken through, synthetic biology in the future will become a problem of pure information science and data science.
In the field of biological manufacturing, cell factory design is the development trend in the future. From the current discrete personal laboratory to the centralized laboratory platform, from distributed data to integrated large-scale data production, this highly standardized and high-quality data provides a great possibility for the final evolution to AI driven design.
If we can use large-scale GPA data sets to deeply mine unknown associated genes and their loci that cannot be found by traditional molecular biology methods from the whole genome based on data science methods, it will be possible to bypass the knowledge bottleneck of rational design from the perspective of data learning, and provide a new research paradigm for improving the efficiency of cell factory design and creation.
In addition, because the large-scale GPA data set has a wider search scope (genome-wide), and does not rely on existing knowledge, it will be possible to explore the phenotypic "highland" that previously could not be achieved by rationality / semi rationality, and obtain the next generation of customized cell factories with more efficient production efficiency and superior production performance.
For this platform, Zhang Zhen's team plans not only to provide a single point of service to scientists, but also to gradually implement academic open source to help the data standardization of multi-dimensional synthetic biology. Driven by high-throughput genotype industrial phenotype association map data, AI parses cell factories to create an enabling platform for synthetic biological data.
The platform will promote the upgrading of scientific research by engineering means, assist scientists to realize technological transformation, and connect the upstream and downstream of the industry. When the platform develops to a certain extent, it will accumulate high industrialization potential energy, and there will be a variety of industrialization possibilities in the future.
"After scientists confirm the strains and phenotypes they want to do by raising needs, we will help them do experiments, and then feed back the results to scientists. At the same time, we hope to deposit the data on the platform, and at the same time, we will open up academic contributions to obtain together, and verify its application value. In the future, this will become the core driving force for the design of several industrial phenotypes." Zhang Zhen said.
It is reported that the team has established extensive cooperation with relevant laboratories of key domestic universities, such as Tsinghua University, Tianjin University, East China University of technology, Shanghai Jiaotong University, Jiangnan University, Shanghai Institute of Botany, Chinese Academy of Sciences, etc. At the same time, Zhang Zhen is carrying out technology licensing and industrialization for the platform. At present, he is preparing to set up relevant companies and set up a technology research and development team. Professor Zhang Zhen is the chief scientist.
Zhang Zhen is full of expectations for the future development of the platform. He said: "I believe that having the core resources of data will also master the core information of cell factory design. We hope that such key databases will take root in China and serve China's local scientific research and industry."”
-End-
Article extracted from deeptech deep technology liuyakun
Original link:https://mp.weixin.qq.com/s/XzmtEshXYE2EvVehboDjAg








